The Virtue of Viability
Every cytometry experiment should contain a viability dye. It doesn't matter if it's just analysis or if you're sorting.
No ifs, no buts.
Dead cells can negatively impact cytometry experiments. They can also skew your functional assays and even your everyday cell culture. Unlike some other experimental and cell handling procedures, the impact of cellular death is easy to quantify in cytometry assays, and as such should be factored into all your experiments.
The scope of this article will only deal with “cellular death” in the context of membrane permeability (intact vs compromised cell membranes) and doesn’t look at issues such as cellular functionality or reproductive capacity, that can also be considered important when assessing sample viability. For example, a cell may have an intact membrane, but be functionally impaired with respect to reproduction and/or differentiation. This article might also be considered “Kingdomist” in that it will predominantly address staining of Eukaryotic (and more specifically mammalian) cells. Remind me to come back and address the issue in relation to Prokaryotes and other sample types one day.
3 Reasons Why
The specific problems associated with dead cells in your cytometry experiments can lead to inaccurate data for 3 main reasons:
- as cells die their membranes become permeable leading to the non-specific uptake of the antibodies and dyes used in your experiment, resulting in false positive results. In the case of rare event analysis in particular, this can result in significant overestimation, or even the identification of immunophenotypic profiles that don’t exist in viable cell populations
- cells stained with cytoplasmic dyes (such as GFP, CFSE or Calcein) will appear to drop expression of these proteins as their membrane permeability increases, resulting in the release of the dye from the cell, and therefore false negative results
- dead and dying cells often become more autofluorescent, which can mask or mimic dim or low antigen expression. In addition, antibodies will be non-specifically caught up by the folds and wrinkles of dying/dead cells, and the exposure of intraceullar contents leads to greater non-specific binding opportunities for antibodies, leading to false postive data
These examples only address the issue of altered staining patterns from dead cells, they don’t deal with the associated issues that arise with the release of the cytoplasmic contents and DNA. DNA is a wonderful natural superglue and if not treated with DNAse, will lead to cell clumping - something that is not ideal when trying to maintain a single cell suspension!
Physically removing dead cells from the tube is very difficult, if not impossible, but it is easy to remove them from the data by the addition of dyes or probes that take advantage of the property of membrane permeability. These dyes come in two main classes - classic DNA binding dyes and protein binding dyes.
Classic DNA-binding Dyes
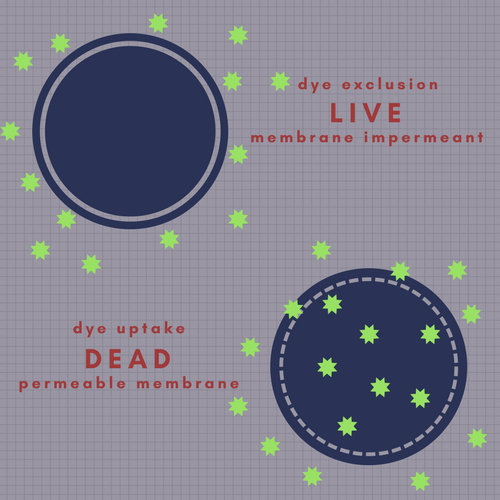
Ask anyone to name a viability dye for you and chances are they they will say PI or 7-AAD. These two reagents have had, and will continue to have a long and illustrious career in flow cytometry applications. They are membrane impermeant, therefore they only stain cells with compromised membranes, while cells with an intact membrane exclude them. They are not the only dyes in this category: other dyes include DAPI, the cyanine dimer family (e.g. TOTO etc), the cyanine monomer family (e.g. TO-PRO etc), SYTOX Nucleic Acid Stains, and one I am particularly fond of for sentimental reasons, Hydroxystilbamidine (FluoroGold).
When using membrane impermeant DNA binding dyes, cells with an intact membrane will exclude the dye and remain negative for fluorescence. Cells with a compromised membrane will take up dye and show positive fluorescence.
Events in this data set were only gated on classical FSC vs SSC parameters, and 2 distinct lymphocyte-like populations are seen.
When a viability dye is added (in this case TO-PRO-3), it becomes apparent that one of these populations is made up of membrane compromised cells as the left population disappears when excluding cells based on dye uptake. Furthermore, downstream gating shows that when the dead cells are excluded, the percentage of CD3+ cells drops markedly. The viability dye has eliminated a significant proportion of false positive events.
These dyes are predominantly inexpensive dyes, and a small amount goes a long way (during my PhD research I used less than 30% of a tube of FluoroGold, and believe me, thousands of tubes were labelled). In addition, the staining process is rapid and easy, and can be done as late as 5 minutes prior to acquisition.
The downside to these dyes is that they are not compatible with assays that require fixation or permeabilisation and the dyes typically have a broad emission spectrum (and sometimes are excited by more than one laser on your cytometer) which can create significant spectral overlap concerns in multi-colour applications.
Propidium Iodide is a very useful dye, particularly in less fluormetrically complex assays, but in multicolour assays the broad emission profile of the dye can result in significant compensation problems across multiple channels.
One solution to this problem comes in the form of protein binding dyes.
Protein Binding Dyes
Protein binding dyes (also called amine dyes) bind to amine groups of cellular proteins rather than DNA. Cells with an intact membrane will present only a few amine groups, resulting in a low level of staining, however, when the dye penetrates the membrane of compromised cells, the dye has access to many more amine proteins found within the cell, and stains with much geater intensity.
Amine viability dyes are a multicolour cytometrists best friend. Not only do they come in a multitude of colours and allow assessment of viability in both fresh and fixed samples, but they typically have a narrower emission spectrum, making it much easier to slot them into a complex panel.
In this example, the Propidium Iodide has been replaced with Live/Dead Fixable Red Stain, which has a notably narrower emission profile, resulting in less spectral spread and improved resolution of data in multicolour panels.
Amine dyes include the Horizon Fixable Viability Stains (from BD), Fixable Viability eFluor dyes and Live/Dead Fixable dyes (from Thermofisher), Viobility dyes (from Miltenyi), Zombie dyes (from Biolegend), Ghost dyes (from Tonbo Biosciences), Live-or-Dye (from Biotium) and VivaFix dyes (from BioRad). Between the vendors there are more than 50 choices - so plenty for you to find a stain to fit your needs.
In order to generate quality, reproducible data, a Viability dye is essential to all cytometry experiments, and with the abundance of choice out there, it is easier than ever before to include this vital tool (forgive the pun) in your cytometry assays.
PS - did you know that you can combine your viability dye in your dump channel in multicolour assays? Just make sure your dump channel antibodies and your viability dye both fluoresce in the same channel and be sure to compensate properly